Review: Chime 2.0

Product Information
Published by: MDL Information Systems, Inc.
Web: http://www.mdli.com/download/chime
System Requirements
PowerPC required
System 7.5 or later
Chime supports Netscape Navigator 3.04 and Netscape Communicator 4.05 and later
Note: Chime does not support Microsoft Internet Explorer on Macintosh
This is not an ordinary software review. The reason is that the software we are talking about is free and of little interest for the most readers. A lot of the popularity of this software is due to the engagement of one man. This may sound a little bit like the Linux story, but it is quite different. The software I am talking about is Chime, a small plug-in for Netscape Navigator that enables the browser to display PDB-files. The man I am talking about is Eric Martz, Professor of Immunology at the Department of Microbiology of the University of Massachusetts in Amherst, MA.
Although not the developer of Chime (which is done by MDL’s staff), his name and work is deeply connected with Chime and its offline predecessor RasMol. He maintains a Website where he features RasMol and Chime and gives a lot of supplementary information for the use of Chime in both teaching and presenting molecules with links to rescoures and FAQs. He also hosts a mailing list dealing with Chime and its functions. Besides of all that, he offers templates for tutorials and full-blown Web-based JavaScript applications using Chime.
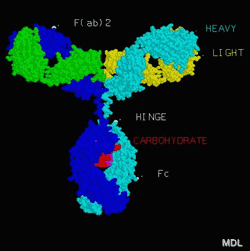
For those who do not deal with molecules and their structures or didn’t come across Chime or RasMol, I want to explain what I am talking about. As I mentioned above Chime is an helper app that enables Netscape browsers to display PDB-files (besides other ones, which I won’t mention). PDB is the format of molecular structures stored in the Protein Databank in Brookhaven. The database contains the structural data of proteins that have been solved using x-ray crystallography. Chime enables your browser to read these PDB data and display them as 3-dimensional models of the molecules.
Of much more interest—especially for teachers—is the fact that Chime is scriptable. And that’s the point where Eric Martz comes back into play. On his homepage and his mailing list he provides a lot of useful information how to write scripts for Chime, to use it in presentations and in teaching. By using his templates, it is a snap to build one’s own presentations of antibodies, hemoglobin, DNA, or whatever. If you don’t want to do it yourself, you also can download the tutorials from his Website and use them to present it to your students. Students themselves may use Chime to explore the world of molecules.
There are two reasons I want to give only a small overview of some basic functions of Chime. First, Chime has a lot of useful features and listing them all would take too long. Second, there is no comprehensive documentation of Chime’s capabilities available from MDL. You have to rely on Eric Martz’ homepage and MDL’s website to get an impression of what you can do with this little piece of software.
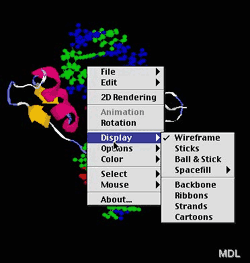
The most basic capabilities of Chime are rotating, dragging, and zooming molecules by moving the mouse. In addition, you can press the command key for dragging, the shift key for zooming, and command-shift for rotating around the z-axis.
The next step is using one of the many features you can find in the pop-up menu. For the case of proteins, you have a variety of display options. You may choose “Spacefill” to see all atoms as balls with Van-der-Waals-radii, or “Wireframe” to see only the covalent bonds between each atom as a fine line. If you want to learn more about the secondary structure you may select “Ribbons” or “Cartoons”. Then only the backbone of the protein and the components of secondary structure like helices, sheets, and loops are drawn.
There are also different options for color-coding proteins. “Structure” colors sheets yellow, helices pink, and loops blue. “Chain” color-codes each chain of a protein. Another feature is the mouse-click option. By selecting “Distance” and clicking on two atoms, the distance between these two atoms is displayed in the status bar of the browser. Another nice tool is the copy menu. “Copy script” copies the script in the clipboard that ran when chime loaded the molecule. The simple “copy” command places a copy of the current display as PICT file in the clipboard. It can then be imported into a word processor or a graphic program for further use.
This is only a small selection of all the features. The best tip for all who want to know more about Chime and its capabilities is to visit the homepage of Eric Martz. The Chime-Plug-in is available for free from MDL’s Website at http://www.mdli.com.
![]() Copyright © 1999 Jens Grabenstein, Jens.Grabenstein@surf24.de. Reviewing in ATPM is open to anyone.
Contact reviews@atpm.com for more information.
Copyright © 1999 Jens Grabenstein, Jens.Grabenstein@surf24.de. Reviewing in ATPM is open to anyone.
Contact reviews@atpm.com for more information.
Reader Comments (2)
Add A Comment